This paper was a long time coming and I am very glad it is finally, officially out (although the resources it describes have been stealthily popular for some time already). The original idea is what brought me to the National Centre for Biotechnology Education at the University of Reading back in 2012, where, along with Dean Madden and John Schollar, we came up with the idea of using some synthetic biology approaches to develop a simple and extensible system of parts and plasmids to enable assembly and characterisation of them in undergaduate settings (check out our poster from SB6 in London). (Later we developed it further, to make it work in secondary schools and six forms, the primary market for NCBE.) We quickly made some progress (see the bacterial transformation kit availabe in NCBE since 2014) and put the protocols for the manipulation of the parts online (the website is no longer available).
Then I started my independent position at Huddersfield and soon afterwards Dean suddenly died; John retired shortly after than and by the time I got around to finishing these resources, it was 2017. It was thanks to two of my project and placement students at Huddersfield, Abbie Williams and Alex Siddall, who took the project up and developed the plasmids and parts to the point where they could be deposited in Addgene for all to use.
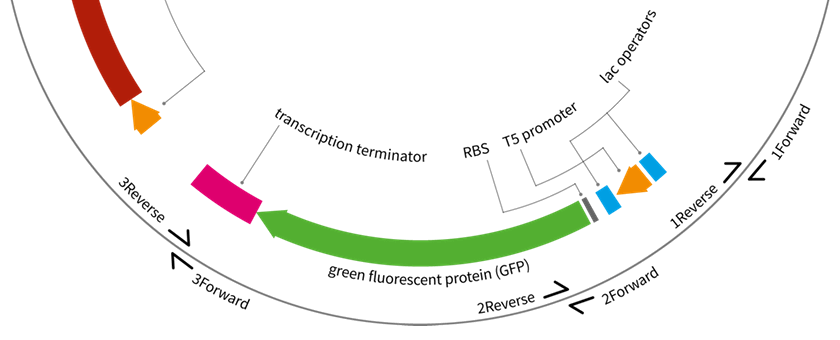
Jason Sanders, another student at Huddersfield, also prepared the graphical materials - plasmid maps - which we hope will be useful for educators to prepare their own assembly diagrams. They are excellent, if I say so myself ;-).
Check them all out and enjoy: Unigems: plasmids and parts to facilitate teaching on assembly, gene expression control and logic in E. coli.
Reuse
Citation
@online{bryk2023,
author = {Bryk, Jarek},
title = {UNIGEMS Paper Is Published},
date = {2023-10-02},
url = {https://bryklab.net/posts/2023-10-02-unigems-paper-is-published/},
langid = {en}
}